After reviewing a diverse collection of 23 submissions, an expert jury has selected a total of six winners—two first prizes of €10,000 each and four second prizes of €3,500 each—in the 2018 GBIF Ebbe Nielsen Challenge. The solutions and innovations devised by the winners—and, indeed, all the entrants—reflect and address topics of wide-ranging interest and concern in the biodiversity informatics community, like linked open data, data mobilization, and data quality and interoperability.
The Challenge is an annual incentive prize that honours the memory of Dr Ebbe Schmidt Nielsen, an inspirational leader in the fields of biosystematics and biodiversity informatics and one of the principal founders of GBIF.
The open-ended nature of this year’s Challenge succeeded in drawing wider geographic engagement, with the 23 entries enlisting participants from two dozen countries. The six winning teams represent eight countries, including two—Sri Lanka and Lithuania—that are not yet formal GBIF members. The complete list of winners, in alphabetical order, is below:
First prize
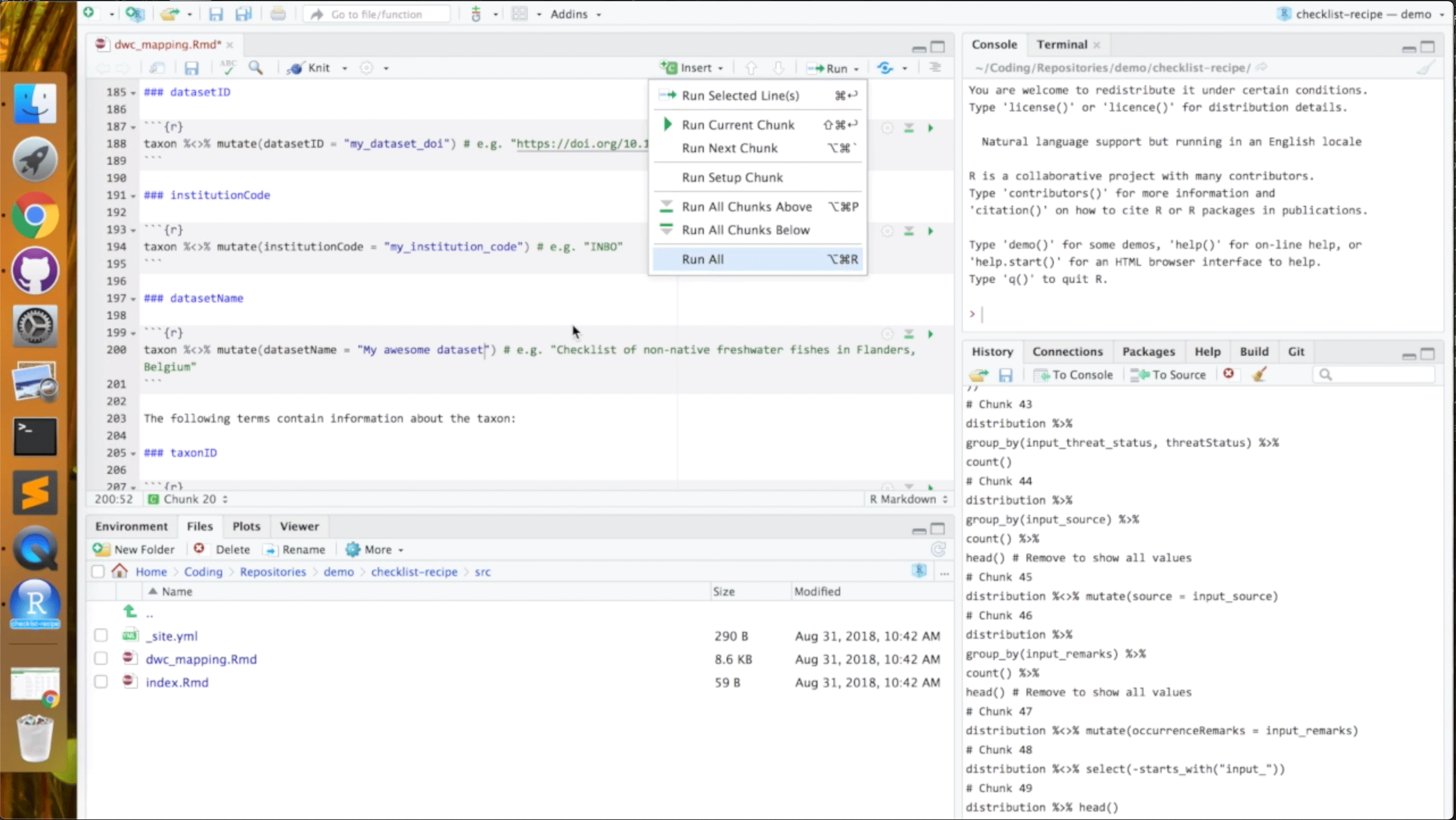
Checklist recipe: a template for reproducible standardization of species checklist data
Lien Reyserhove, Damiano Oldoni and Peter Desmet, INBO: Research Institute for Nature and Forest, Belgium
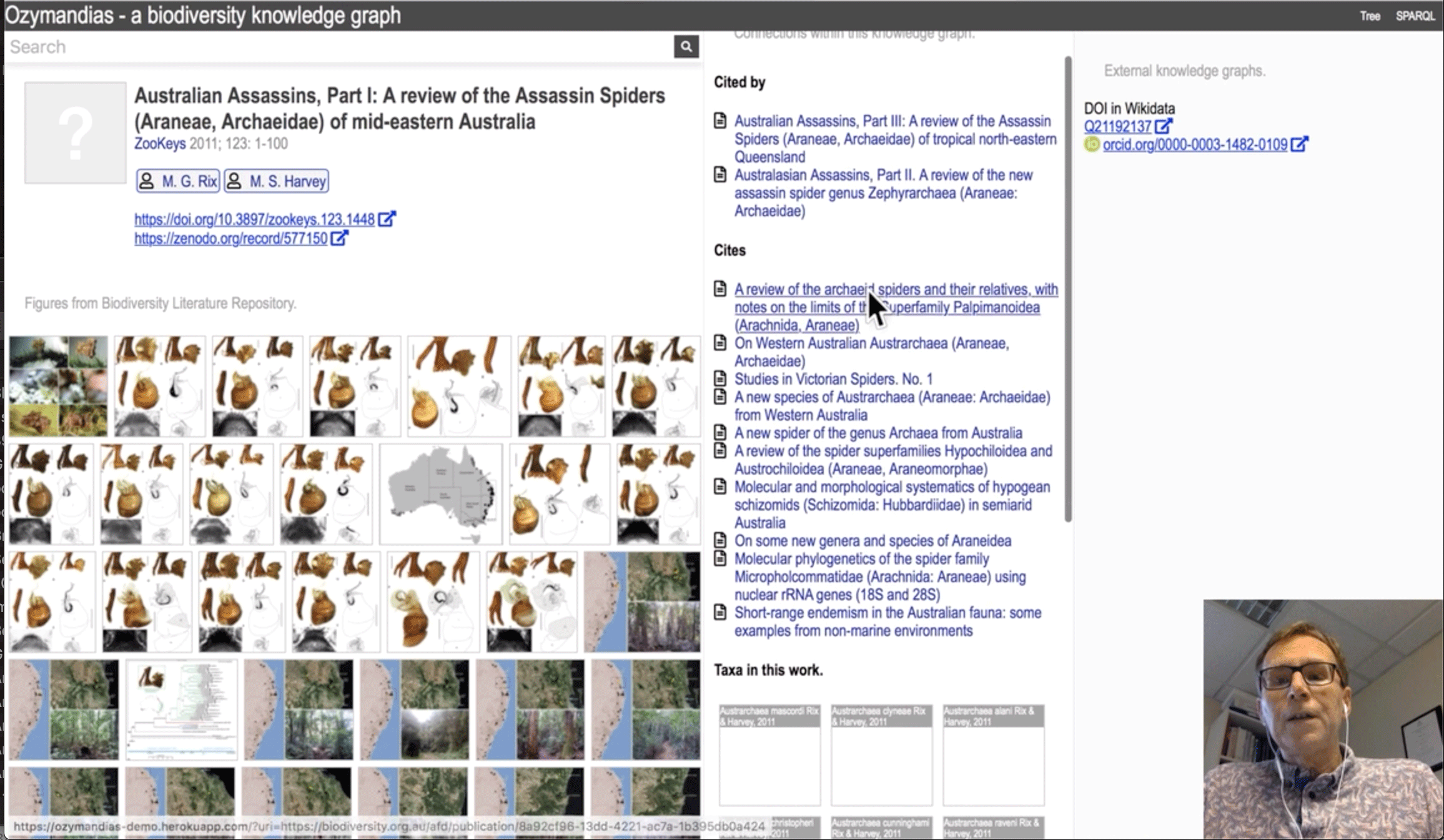
Ozymandias: a biodiversity knowledge graph
Roderic D. M. Page, University of Glasgow, United Kingdom
Second prize
The bdverse
Tomer Gueta and Yohay Carmel, The Technion – Israel Institute of Technology, Israel
Vijay Barve, Florida Museum of Natural History, United States
Thiloshon Nagarajah, Informatics Institute of Technology, Sri Lanka
Povilas Gibas, Vilnius University, Lithuania
Ashwin Agrawal, Indian Institute of Technology (BHU), India
GBIF Issues Explorer
Luis J. Villanueva, Smithsonian Institution, United States
Smart mosquito trap to DwC pipeline
Connor Howington and Samuel Rund, VectorBase.org, United States
Taxonomy Tree Editor
Ashish Singh Tomar, University of Granada, Spain | BIG4
The recipients are share a larger-than-normal share of prize money thanks to an earlier contribution from the Swedish Research Council, which went unawarded in last year's Challenge.
First-prize winners
Checklist recipe
The team of Lien Reyserhove, Damiano Oldoni and Peter Desmet from INBO, the Flemish Research Institute for Nature and Forest, cooked up Checklist recipe, an automated routine that uses the R programming language to generate species checklist datasets. This recipe solved an ongoing need for the Tracking Invasive Alien Species project (TrIAS), which aims to support evidence-based policies on invasive and alien species (IAS).
As the team's submission noted, "Standardizing biodiversity data to Darwin Core can be hard." Their template GitHub repository provides a transparent, open, and repeatable process for mobilizing and publishing thematic species checklists from diverse sources.
Users with a basic knowledge of R can copy—or 'clone'—the repository, customizing the code to fit their needs while transforming source data files into a standardized Darwin Core checklist. The recipe also supports several essential parts of the data processing workflow, including iterative versioning and data validation, mapping, correction and republication.
"TrIAS is an open science project, so we wanted to make the standardization routine for our checklist data open and reproducible as well," said Desmet. "The recipe has considerably streamlined our own work to publish seven checklists on alien species for Belgium. We thought it would be useful to provide a well-documented workflow for others who want to publish this type of data.”
The team is now working to reuse the open checklist data they’ve already published to create a reproducible national list of alien species in Belgium as part of the Global Register of Introduced and Invasive Species.
Ozymandias: a biodiversity knowledge graph
The core concept for Ozymandias has long been a preoccupation of its creator, Roderic D.M. Page, professor of taxonomy at Glasgow University and former chair of the GBIF Science Committee. Using shared identifiers to crosslink openly available information on taxa, journals, publications and people, this 'biodiversity knowledge graph' reveals the connections between researchers, research outputs and the data their work supports while, in Page's words, moving 'beyond tables to thinking in terms of connected networks of relationships.'
The initial prototype for Ozymandias submitted for the Challenge combines a classification of animals from the Atlas of Living Australia with data on taxonomic names and publications from the Australian Faunal Directory. Common identifiers like DOIs (Digital Object Identifiers) for articles, LSIDs (Life Science Identifiers) for taxonomic names, and ORCIDs and WikiData IDs for people trace back to their sources in scientific journals, scanned literature from the Biodiversity Heritage Library, citation events from CrossRef and figures from Plazi's Biodiversity Literature Repository.
Even constrained to the scale of a single continent, Ozymandias enables casual users, students and researchers to explore millions of relationships and use any part of it as the entry point for investigation, focusing on knowledge of a particular species, the range of a researcher's activities or the research output associated with a particular institution.
"Knowledge graphs like Ozymandias have numerous practical applications, such as informing data collection and management policies, for example, by discovering gaps in literature digitization, or uneven representation of content from different institutions," Page notes. "They also provide a critical tool for engaging taxonomic researchers in data curation, because if a researcher has an ORCID, we can discover their list of publications and identify the taxa that they work without having to ask for demonstrations of their expertise."
Page hopes that future iterations of Ozymandias will add links to specimen and sequence data while expanding its taxonomic scope and geographic range.
Second-prize winners
The bdverse
The aspirations of this Challenge submission are high: creating nothing less than a general framework to bridges between all biodiversity data users and the R ecosystem while promoting the standards and methods of the Biodiversity Informatics community. Led by Tomer Gueta and Yohay Carmel at The Technion – Israel Institute of Technology in Haifa, the competition's most diverse international team assembled six unique R packages in a hierarchal structure that tackles distinct functions. From standardizing fields and checking and cleaning issues to preparing visualizations and reports, each package operates on a collection of user-friendly Shiny apps and a complete set of R functions. The "Ebbe" version of the bdverse comes in advance of an official release planned for January 2019. The team now is now building a testing framework with continuous integration and is eager get feedback about bdverse's architecture, its features and its user interface.
GBIF Issues Explorer
Improving the quality of previously published data can be time-consuming job, especially for collection managers responsible for large datasets. The Challenge entry from Luis Villanueva of the Smithsonian Institution seeks to simplify the process of reviewing issues flagged in individual occurrence records. The application provides a web interface that enables users to browse issues by dataset, highlighting patterns, trends and other relevant information. Meanwhile, the tool displays inline images, key columns (like coordinates, location, and country), links to the collection database and a map to help collection managers sort out where the problems reside and how to fix them in subsequent updates. Researchers can also use Issues Explorer to check datasets and gauge their suitability prior to running their analyses.
Smart mosquito trap to DwC pipeline
Can smart sensors automate the capture of standardized time-series species occurrence data? For their Challenge entry, Connor Howington and Samuel Rund from VectorBase.org have developed a workflow for processing real-time time abundance data gathered by commercially available devices for remote mosquito monitoring. The routine starts with pulling data from the BG-Counter’s proprietary API, condensing, cleaning and processing it to align with the open Darwin Core format. Their proof of concept represents a timely first step in preparing standardized open data from smart traps, given that next-generation sensors in development analyze wing-beat frequency to provide accurate species-level identifications.
Housed at the University of Notre Dame, VectorBase.org is one of six Bioinformatics Resource Centers for Infectious Diseases supported by the U.S. National Institute of Allergy and Infectious Diseases, part of the National Institutes of Health.
Taxonomy Tree Editor
This Challenge entry from Ashish Singh Tomar of the University of Granada offers a prototype web application for visualizing taxonomy trees, allowing taxonomists to edit and to compare a branching graph of evolutionary relationships against the GBIF backbone taxonomy. Tomar has developed the tool as a postdoc in BIG4, a global consortium training a new generation of systematic entomologists—but taxonomists anywhere could benefit from being able to drag-and-drop missing nodes from a reference taxonomic tree into their working taxonomy.
Jury for 2018 Ebbe Nielsen Challenge
- Greg Riccardi, iDigBio
- Jo Judge, UK National Biodiversity Network
- Marie Elise Lecoq, GBIF France
- Eduardo Dalcin, Rio de Janeiro Botanical Garden Research Institute
- Dag Endresen, GBIF Norway
- Ana C.M. Malhado, Universidade Federal de Alagoas
- Anabela Plos, GBIF Argentina | Museo Argentino de Ciencias Naturales
- Anders G. Finstad, Norwegian University of Science and Technology
- Philippe Grandcolas, Muséum national d'histoire naturelle