The registry is a core component of the architecture responsible for providing the authoritative source of information on GBIF Participants (Nodes), institutions (e.g. data publishers), datasets, networks their interrelationships and the means to identify and access them.
As a distributed network, the registry serves a central coordination mechanism to (e.g.) allow publishers to declare their existence and for data integrating components to discover how to access published datasets and interoperate with the publisher.
Purpose
The role of the registry can be summarised as a component offering:
- An authoritative source of information (metadata) on institutions, datasets, technical services and other key entities as required by registry partners. Due to the nature of the network and tools in use, multiple versions of this information are often available. Where this occurs, the registry aims to provide the most complete representation by merging sources and harmonizing conflicting views where possible. This simplifies consumption to clients by providing a unified view of metadata in a consistent format. Links to external representations available through other formats (such as Ecological Metadata Language are available to clients.
- A source of information on inter-relationships between datasets, institutions and other entities according to the needs of the registry partners. Datasets can be hosted by one party on behalf of another, and might themselves be a superset of other datasets. Modelling of dataset relationships provides an indication of where duplicate content might exist and how to correctly determine the attribute chain for all parties involved in the data management lifecycle.
- A trustworthy identifier assignment (minting) service for institutions and datasets. Identifiers are allocated as Universally Unique IDentifiers (UUID) on first registration, and datasets and downloads are given a Digital Object Identifiers (DOI) for external use.
- An identifier resolution service allowing external clients to submit a known identifier and resolve this to the registry assigned identifier. Thus clients already using Biodiversity Collection Index (BCI) identifiers (or others) can interact with the registry using BCI identifiers. The number of identifier systems recognised is expected to grow continuously as more systems are connected.
- A mechanism to help coordinate distributed system activities by
- providing preferred technical access points where multiple routes exist
- offering stable identifiers for registered entities and
- providing notification services of significant events such as a dataset being registered
- A discovery mechanism for users and machines for
- Registered network entities
- Technical endpoints
- Data definitions (for example, standards) such as the extensions and vocabularies used in the Darwin Core Archive format
- Discovery is provided through indexing of metadata, and through flexible tagging of entities using simple key value pairs of tags, optionally in a restricted namespace. Tagging may be done publicly (no namespace), allowing anyone to make use of the tag, or by maintaining a private collection of tags (private namespace). Private tagging ensures a registry client can define their own terms (vocabulary) for tagging and be assured others cannot make changes to their tags. An example of a public tag on a dataset might assert that
basisOfRecord = Specimenwhich might be useful to many whereas a private tag in a scratchpad namespace might assert thatscratchpads.eu:activity = highwhich is specific to the namespace owner and cannot be changed by others. Please note tagging is done exclusively through the web services API.
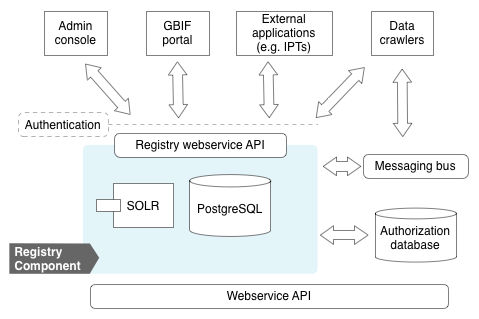
Component Architecture
At the heart of the registry is a PostgreSQL database (view schema) accessed exclusively through a web services API. To enable faceted search, an Apache SOLR index is maintained and exposed in the API. The registry emits messages to a messaging bus (RabbitMQ) to enable components to subscribe to significant events, such as a newly registered dataset to be ingested.
Evolution
A registry has existed as a key component of the infrastructure since the conception of the GBIF network. Originally built on the open industry business registry known as UDDI, the registry recorded institutions, their contact information and their hosted technical services.
When the GBIF Data Portal was designed in 2007, data harvesting tools interacted with the registry to locate endpoints and crawl the network, building the necessary search indexes to drive the portal services. In 2009 when the Integrated Publishing Toolkit was released, the registry incorporated a RESTful web service interface enabling automatic registration of published datasets. This was known as the Global Biodiversity Resources Discovery System (GBRDS).
Today the registry has evolved to provide a RESTful web service API, search services and means to discover data standards and vocabularies used during the data publication process.
For details on other components of the GBIF architecture, please refer to the list at the top of this page.