The network that publishes occurrence records through GBIF spans hundreds of publishing institutions worldwide. Data holders manage content in either spreadsheets or databases and then use specific publishing tools to expose those data for querying and access over the internet. The existence of the dataset and the technical protocols required to access the data are entered into the GBIF registry.
Aggregators, such as GBIF.org and national GBIF data portals, download datasets and build sophisticated indices to allow users to efficiently search and access content across datasets.
This page briefly describes the architecture and operations performed by GBIF when retrieving and indexing occurrence data for user search and download.
Component architecture
The architecture is designed to be an efficient distributed data ingestion and processing system, with decoupling of components. The primary goals of the architecture design were to ensure rapid processing of data to reduce latencies between publisher changes and global discovery, and to be flexible enough that components can be swapped out or updated as needed. The architecture is best explained with 2 example use cases.
- By describing the sequential set of steps that occur when a new dataset is registered into the GBIF registry. This use case was chosen since a newly registered dataset triggers action on all components of the architecture.
- By describing the process that occurs when a user searches and chooses to download content.
The following illustrates the core components of the occurrence architecture which will be explained as we progress through the examples. Core to all of this is the distributed Apache Hadoop cluster, which provides distributed redundant copies of content and parallelized processing capabilities.
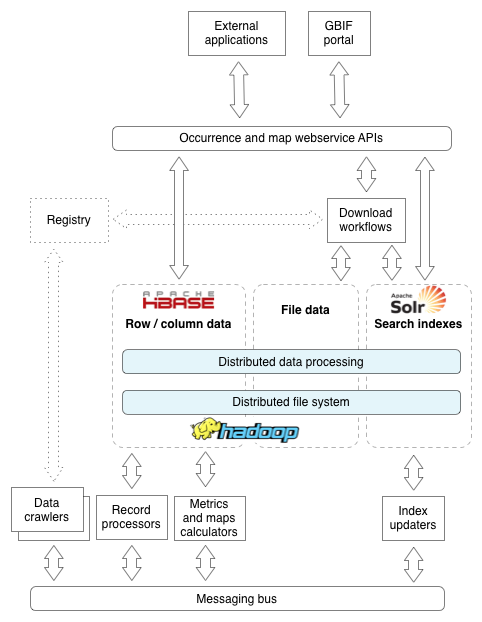
Use case: A newly registered dataset
- The dataset is registered in the registry. The technical access point information is associated with the dataset, including the URL and protocol required to access it.
- The registry emits a message onto the shared messaging bus (RabbitMQ) that a dataset is ready to be crawled. Note that the crawl scheduling component might also emit this message when a dataset is ready to be re-crawled which happens periodically, typically every seven days
The dataset will be entered on the data ingestion monitor.
Crawling: This is the process where the dataset is downloaded, either as a single download for Darwin Core Archives, or as many successive requests for XML-based datasets. - There are many crawlers that can run concurrently, and are controlled by the crawler coordinator. The coordinator receives the message from the registry and will manage crawl resources to initiate the crawl. At this point, locks (Apache ZooKeeper) are taken out to ensure that concurrent crawlers do not over-eagerly crawl an endpoint. Additionally, shared counters will be configured, which will be updated at every subsequent stage of processing to allow for overall monitoring, and the ability to determine when a crawl is complete across all components.
- Some of the data protocols GBIF support require paging (TAPIR, BioCAsE and others), while others support a single HTTP request (Darwin Core Archives and metadata-only datasets). In both cases, for each artifact retrieved (a page or the full dataset) the crawler downloads the content, persists it to disk and emits a message declaring it is ready to process. Shared counters are maintained about the progress. For Darwin Core Archives, validation will occur to ensure the archive is suitable for further processing
- For each artifact, a processor will extract single occurrence records, emitting a message for each record harvested. This is received and a processor inspects the record for its identity, either
dwc:occurrenceIDor using thedwc:institutionCode,dwcCollectionCodeanddwc:catalogNumber. In this case the records are all new and the fragment for the record will be inserted into Apache HBase. This is the raw content as harvested, so might be a fragment of XML for some protocols. A message is emitted and counters updated. - A processor will receive the message and the raw view of the record is normalized into separate fields of Darwin Core terms with as little interpretation as possible. A message is emitted declaring the record is normalized and a processor will start interpreting content and applying quality control. This includes:
- Ensuring all basis of record values align to a vocabulary
- Ensuring all coordinates look sensible compared to the other fields present
- Making inferences of (for example) country, where coordinates exist but no country is stated
- Aligning the record to the GBIF Backbone taxonomy using the name lookup web service
Once processing is complete, a message is emitted to declare the record is finished, and counters are updated. The crawler coordinator monitoring the counters will observe this, and will write the result of the crawl to the registry for future auditing, and reporting on crawling history when it determines from the counters that all messages are processed.
- In order to support real time search, and various views on the portal such as maps and statistics, specific data structures are maintained. The newly created record messages will be observed by Apache SOLR index updaters which will update the index
Use case: User search and download
- The user performs a search in the portal and chooses the download button.
- A download workflow is initiated, which is orchestrated by the Hadoop workflow manager provided by Apache Oozie.
- The workflow will follow the following process:
- A query against Apache SOLR is used to get the count of records to download
- If the result size is below a threshold, a paging process iterates over results from SOLR, retrieving the occurrence identifiers. For each identifier, a "get by key" operation against HBase provides the record details, which are written into the output file.
- If the result size is above the threshold, a Hadoop MapReduce-based process is initiated. Apache Hive is used to provide an "SQL like" query language, which iterates over the HBase content to produce the output file
- Once complete, subsequent workflow stages run which produce the necessary citation and metadata files by using the registry API
- Finally all pieces are converted into a Darwin Core Archive by zipping together, or a compressed CSV file. The Zip format is chosen as it is the most compatible.
For details on other components of the GBIF architecture, please refer to the list at the top of this page.